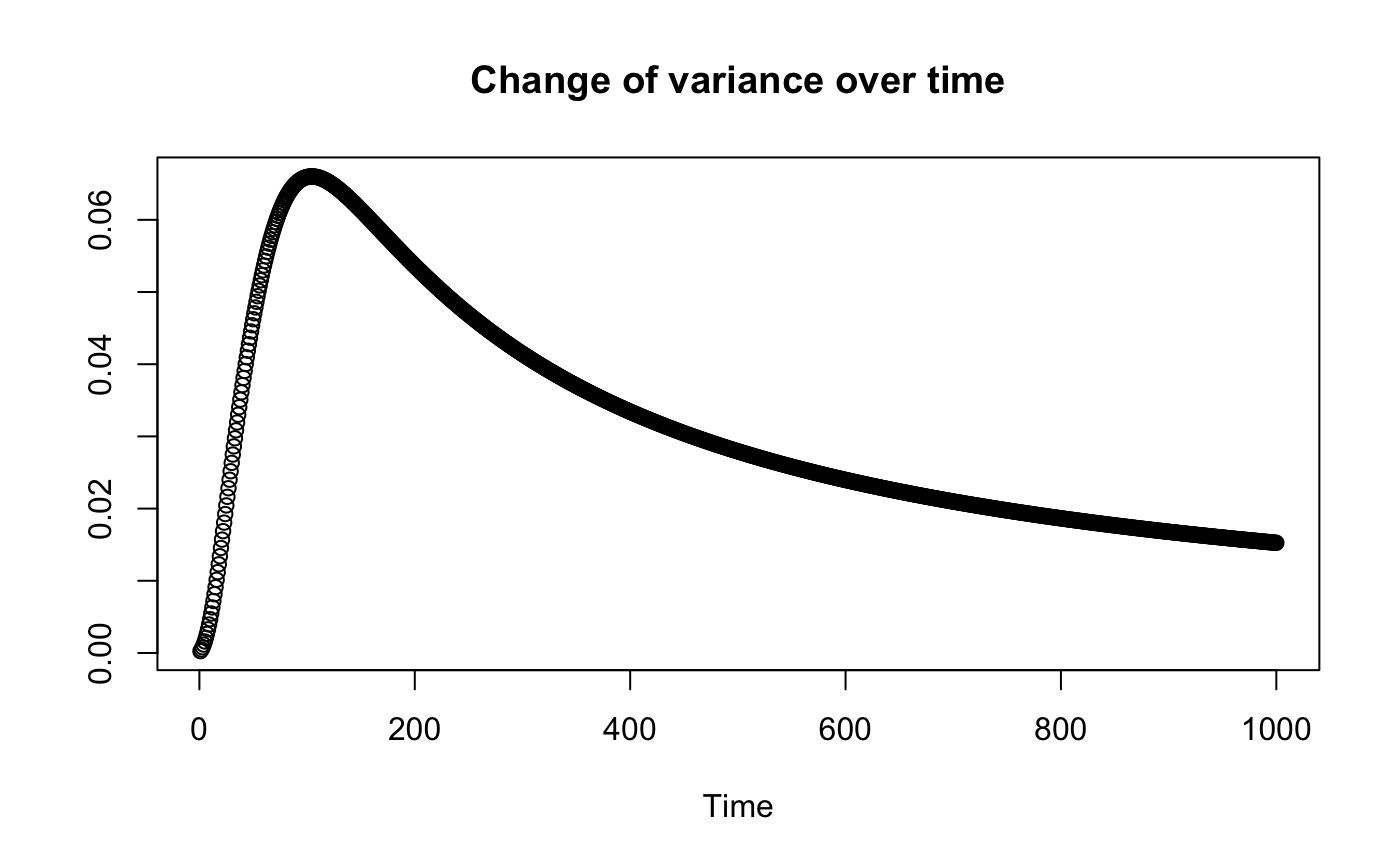
Plot the mean variance across all taxa for an increasing number of time points. If groups are given, compute the mean of beta diversity (assessed with Bray Curtis) across samples for each time point. Groups are interpreted as replicates, such that each group is assumed to cover the same time points in the same order.
varEvol(x, groups = c(), plot = TRUE, sd = FALSE)
Arguments
| x | a community matrix with rows representing taxa and columns time points |
|---|---|
| groups | a vector with group assignments and as many entries as there are samples |
| plot | do the plot |
| sd | use the standard deviation instead of the variance |
Value
a list with variances (or standard deviations if sd is true) and the intersection, slope, pval and adjR2 of the linear regression of variances against time
Examples
#> [1] "Adjusting connectance to 0.02" #> [1] "Initial edge number 400" #> [1] "Initial connectance 1" #> [1] "Number of edges removed 373" #> [1] "Final connectance 0.0184210526315789" #> [1] "Final connectance: 0.0184210526315789"out=varEvol(ts)#> [1] "Change of variance of dissimilarity across replicates" #> [1] "Slope: -4.23252458466131e-05" #> [1] "Adjusted R2: 0.613002261934749" #> [1] "P-value: 0"