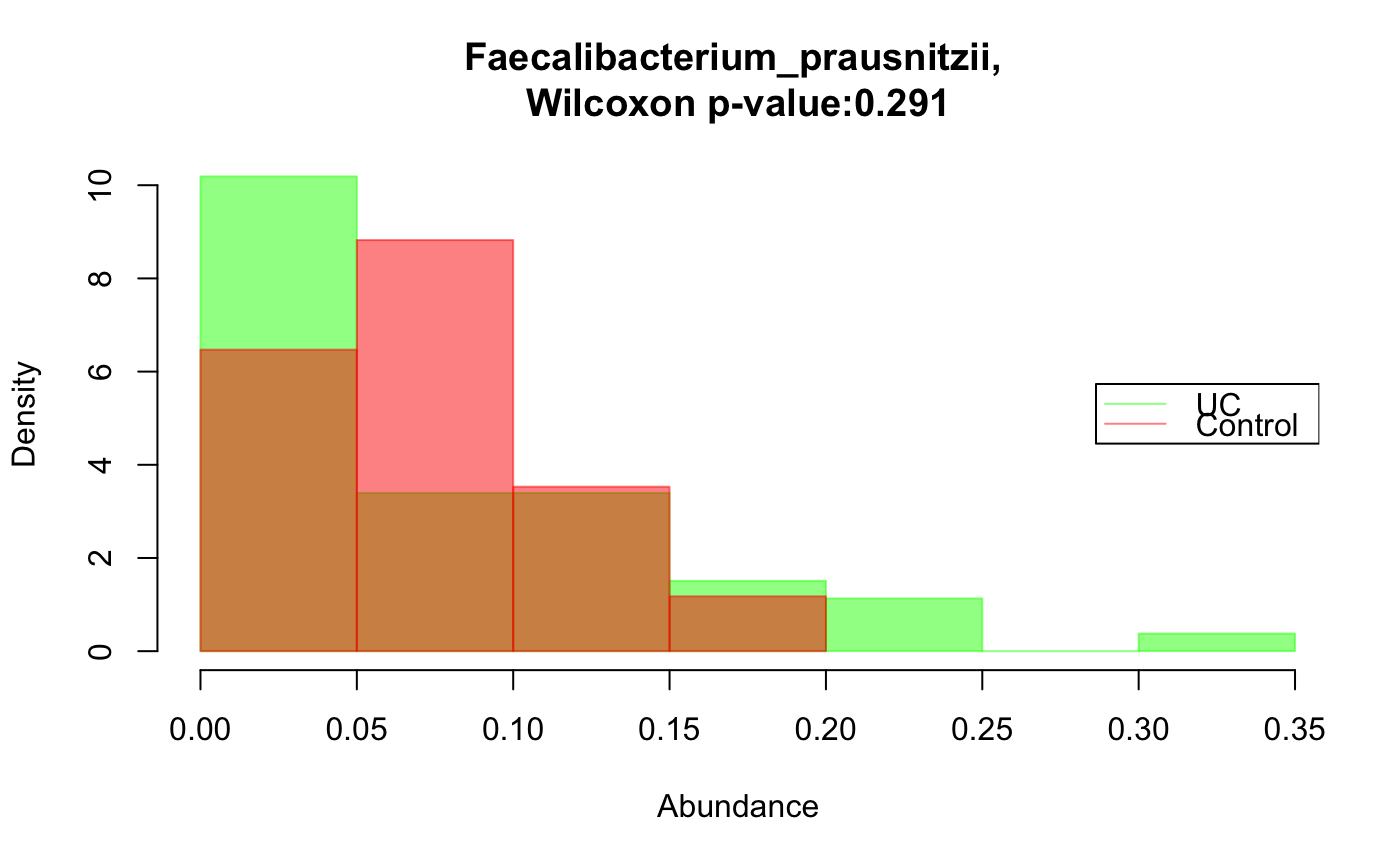
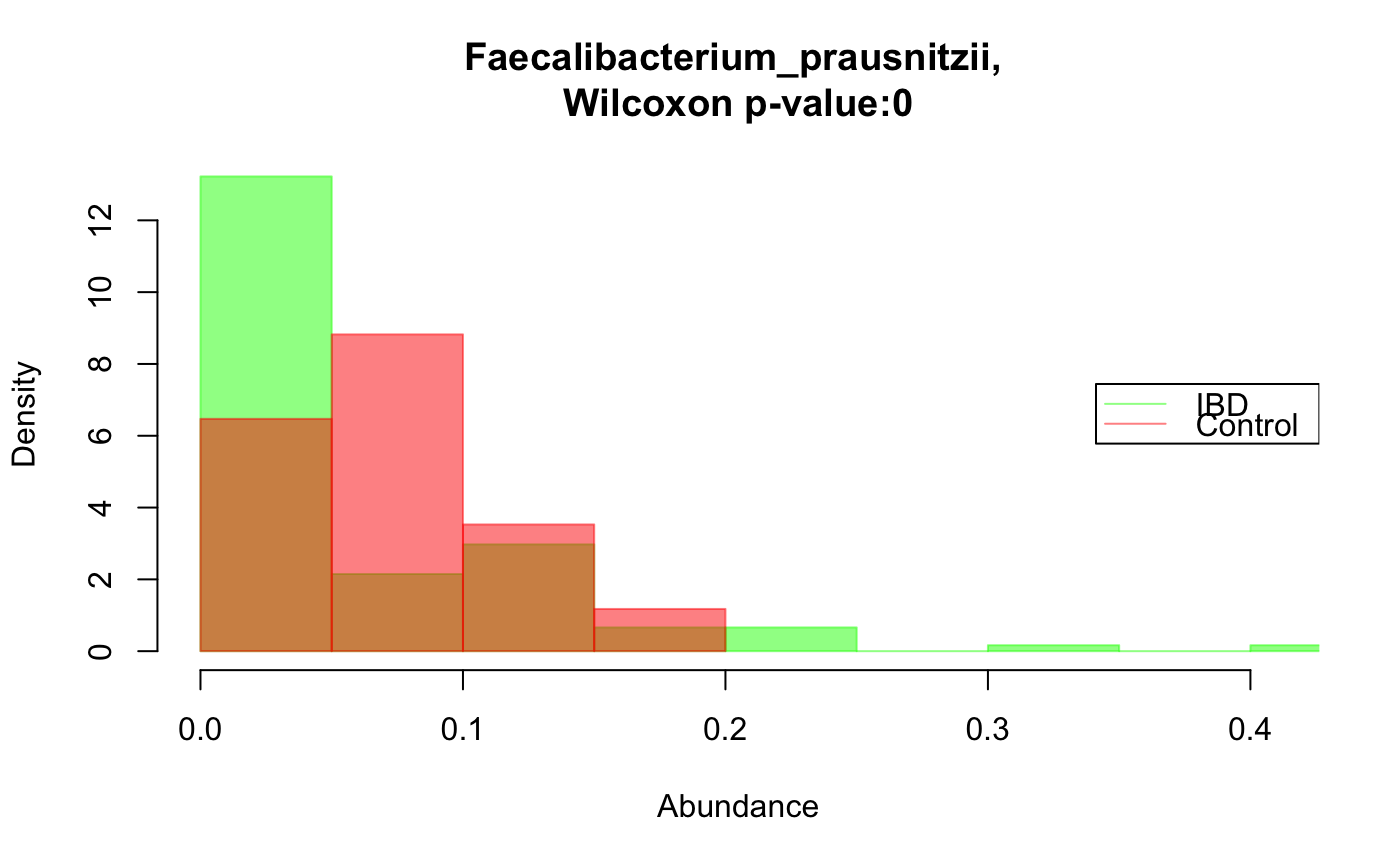
Compare the distribution of a taxon across two groups
compareDistribs.RdPlot taxon abundances as histograms and calculate the unpaired two-sided Wilcoxon p-value.
compareDistribs( x, taxon, groups = c(), group1 = 1, group2 = 2, name1 = as.character(group1), name2 = as.character(group2) )
Arguments
| x | a matrix with taxa as rows and samples as columns |
|---|---|
| taxon | the index or name of a taxon to be compared |
| groups | a vector that provides for each sample its group membership (numeric or character) |
| group1 | the identifier of the first group (numeric or character) |
| group2 | the identifier of the second group (numeric or character) |
| name1 | the name to be displayed for the first group |
| name2 | the name to be displayed for the second group |
Examples
data("ibd_taxa") data("ibd_metadata") groups=as.vector(ibd_metadata$Diagnosis) taxon="Faecalibacterium_prausnitzii" compareDistribs(ibd_taxa,taxon=taxon,groups=groups,group1="UC",group2="Control")groups[groups=="UC"]="IBD" groups[groups=="CD"]="IBD" # Faecalibacterium has a significantly higher relative abundance in the control group compareDistribs(ibd_taxa,taxon=taxon,groups=groups,group1="IBD",group2="Control")